Functional profiling
Key concepts
Assessed profiles:
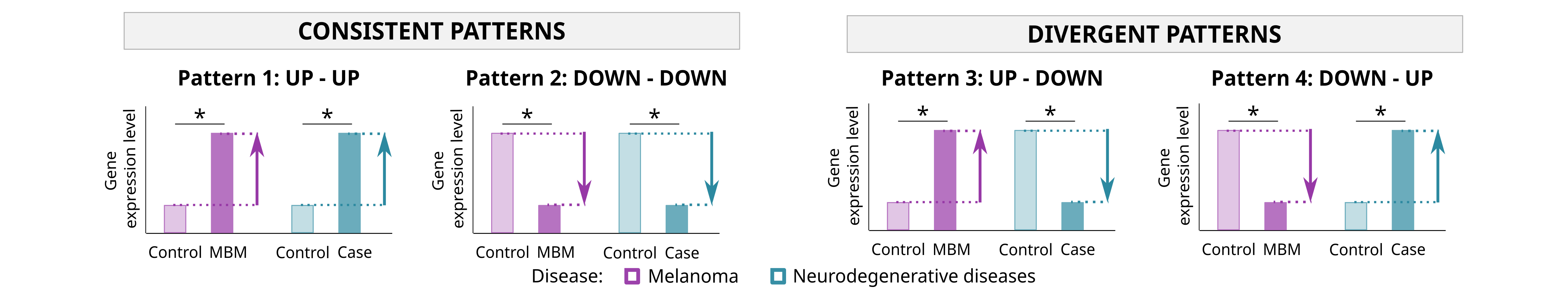
- By pattern: functional analysis of genes presenting the same dysregulation pattern in MBM-3 and in neurodegenerative diseases. Patterns evaluated:
- By neurodegenerative disease: functional analysis of genes altered in MBM-3 and a specific neurodegenerative disease, independently of their dysregulation pattern. Neurodegenerative diseases evaluated:
- Cortex region of Alzheimer’s disease (AD-CT)
- Hippocampus region of Alzheimer’s disease (AD-HP)
- Substantia nigra region of Parkinson’s disease (PD-SN)
Functional results
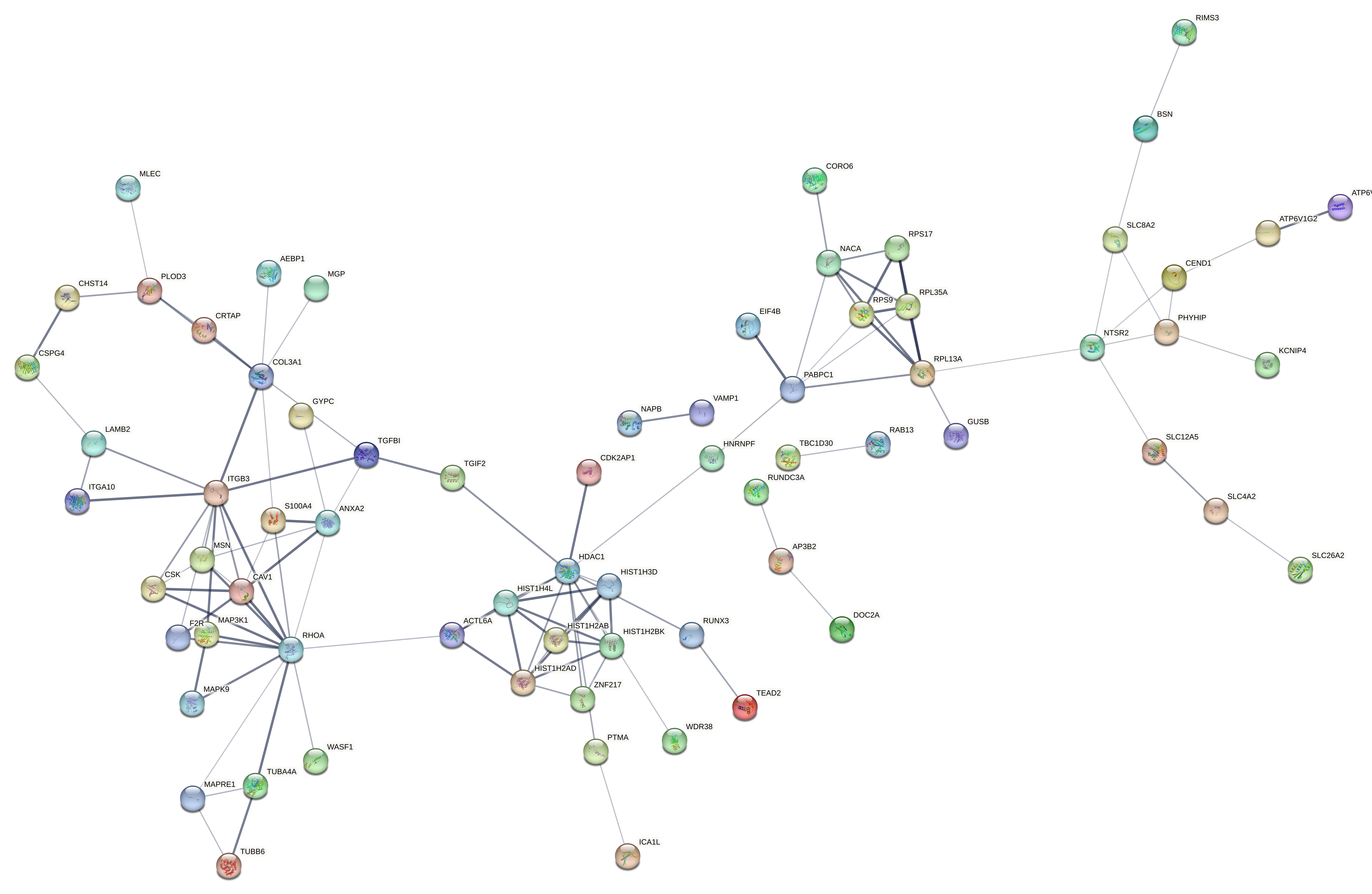
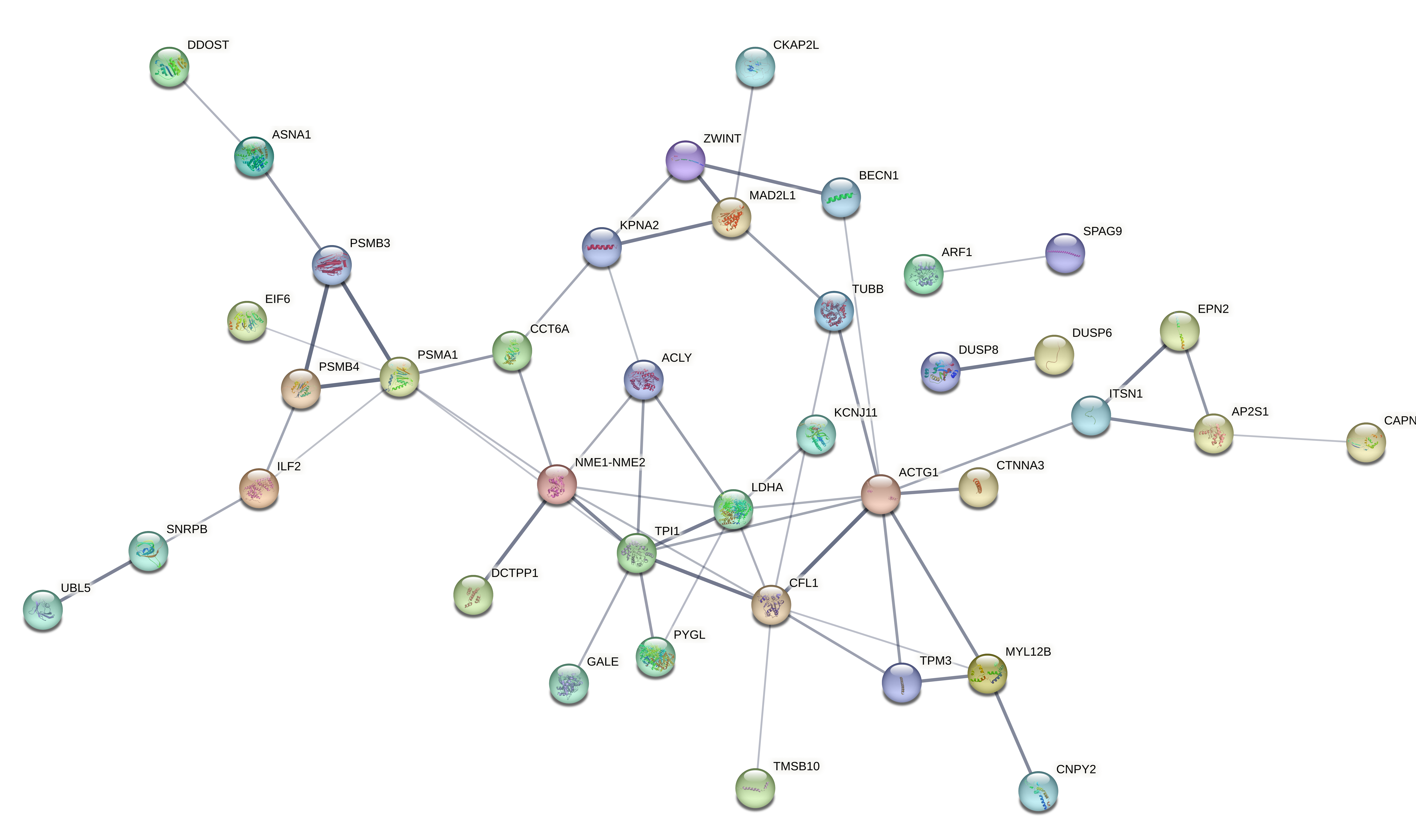
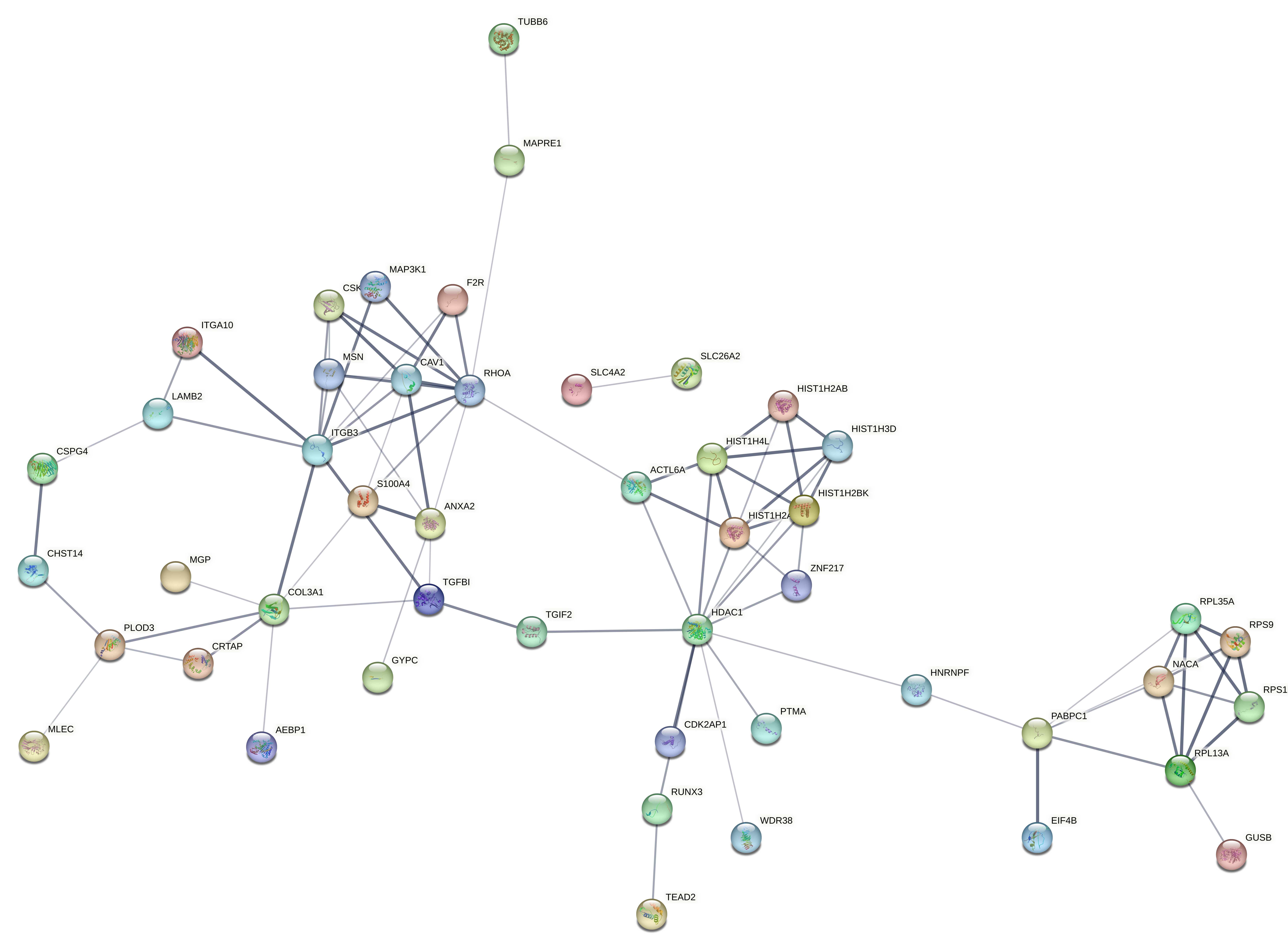
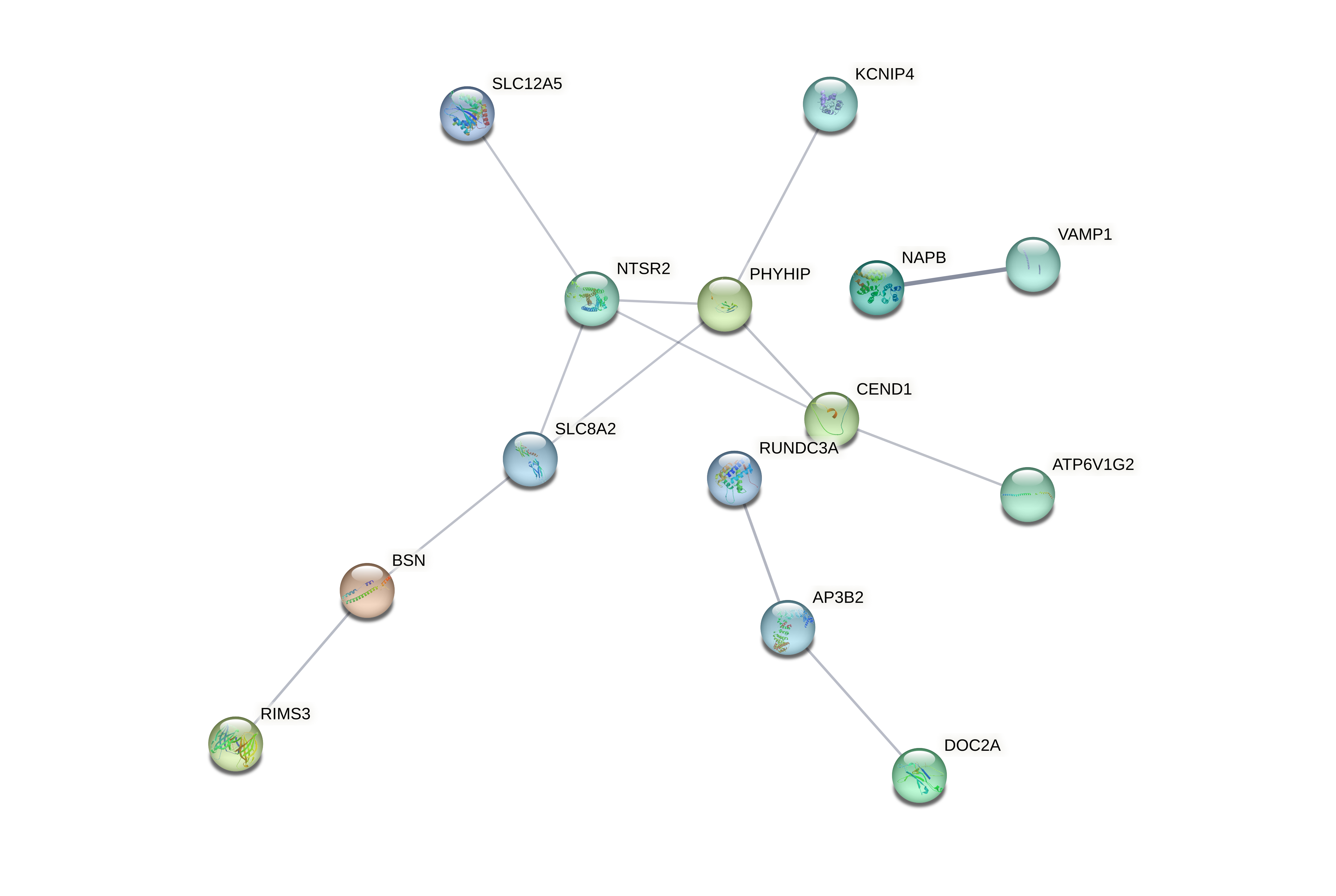
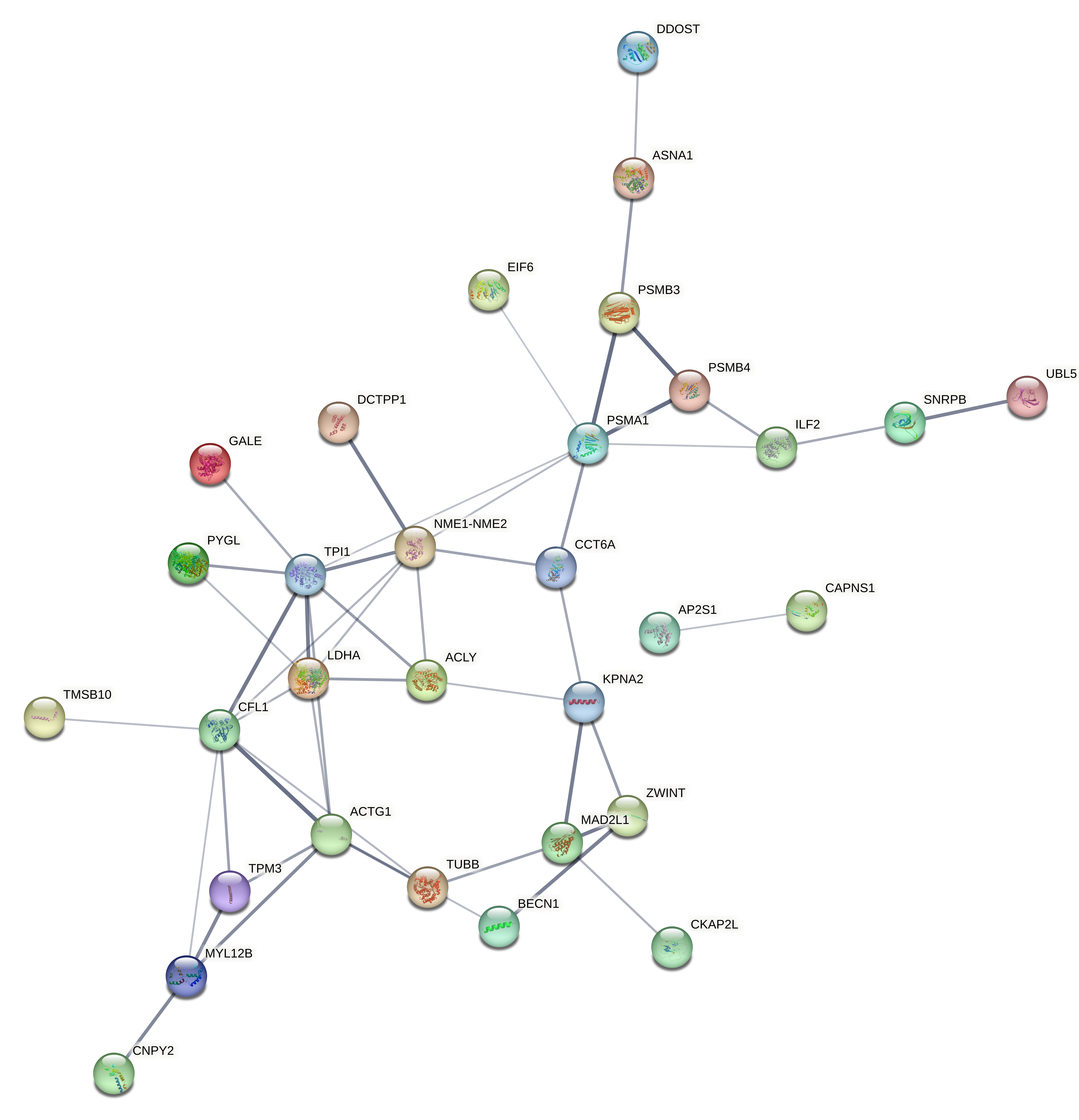
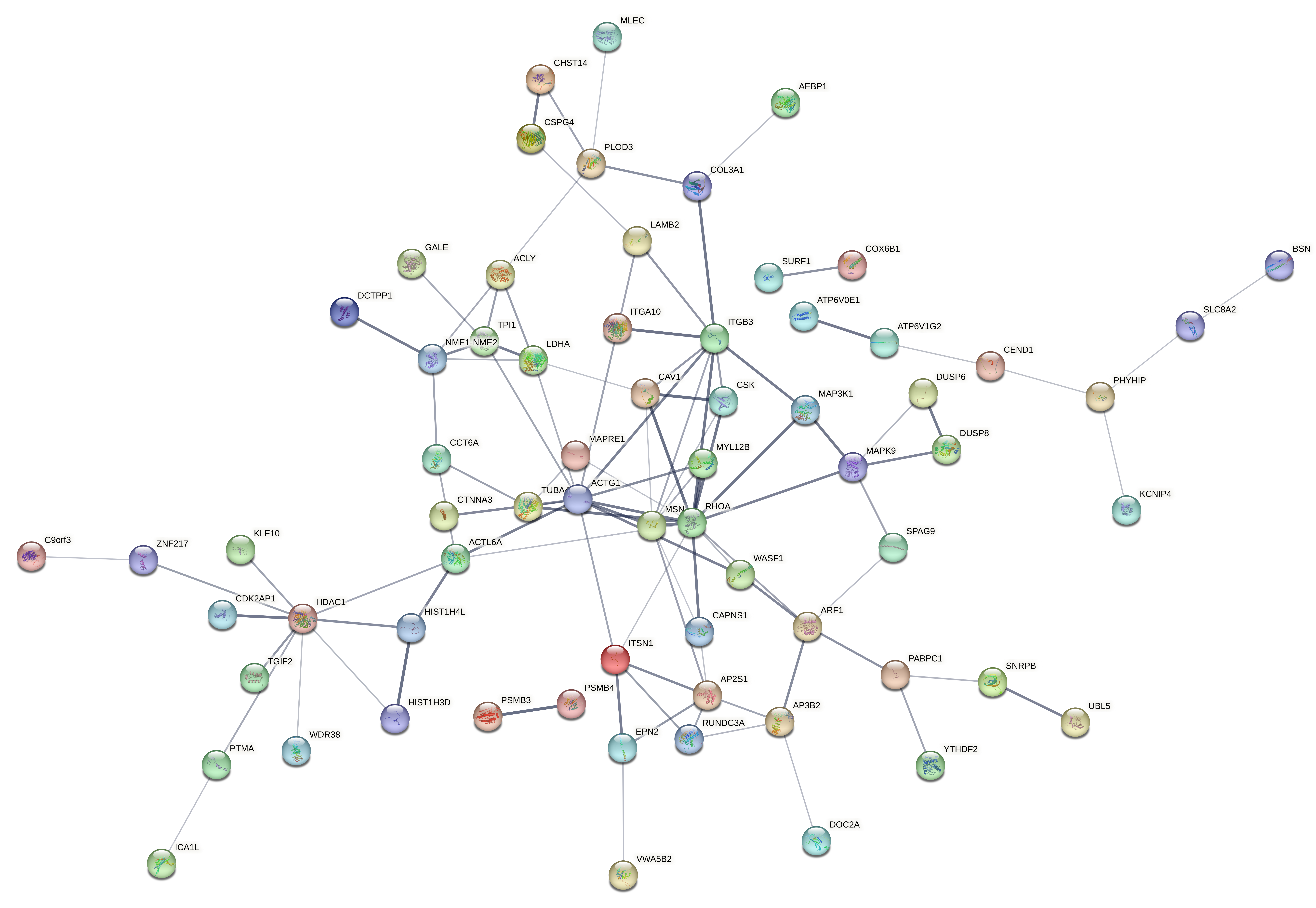
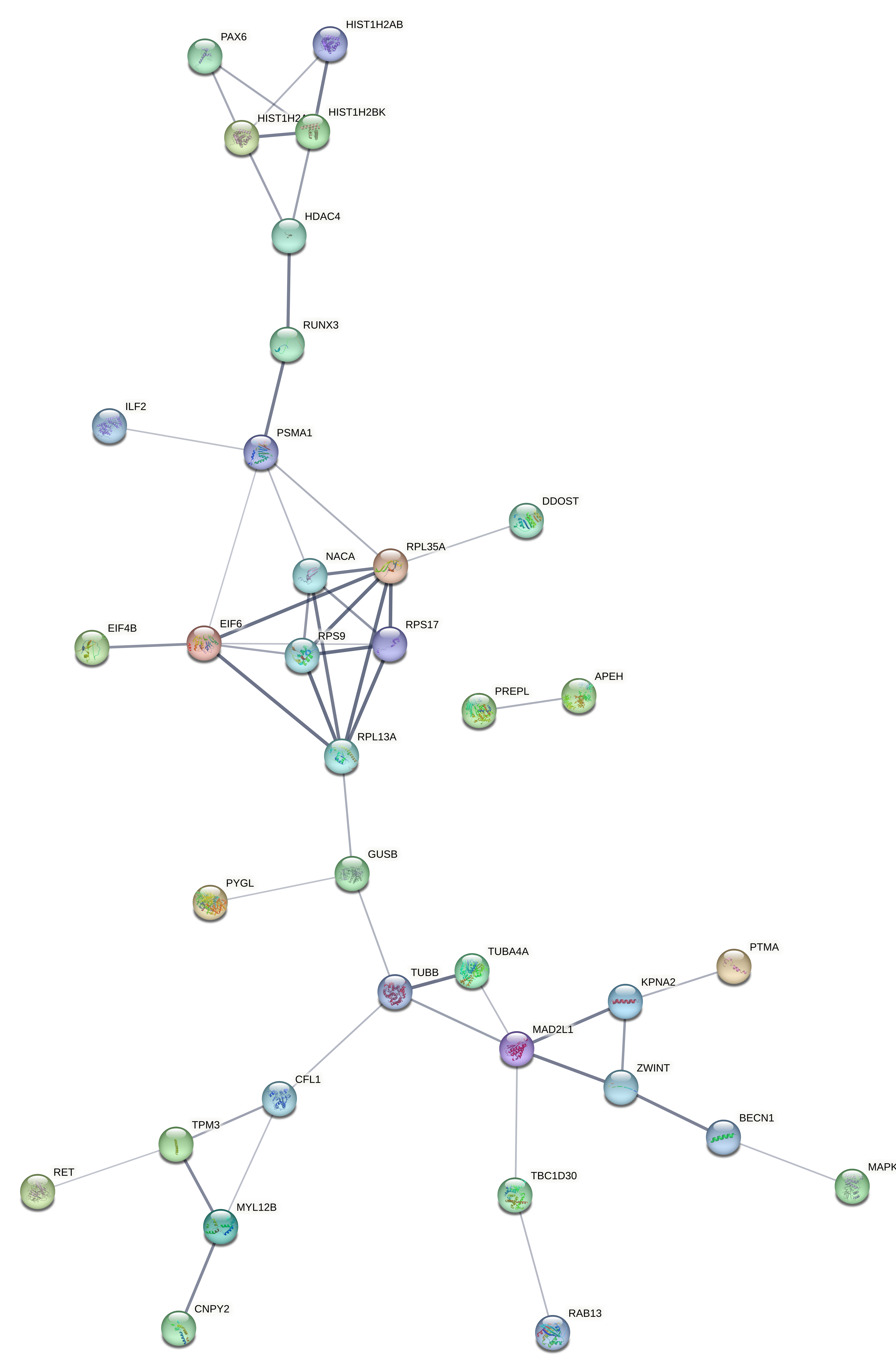
Evaluated genes: Gene list: A2ML1, AAR2, ACBD7, ACLY, ACSBG2, ACTG1, ACTL6A, AEBP1, AIFM3, ANXA2, AP2S1, AP3B2, APEH, ARF1, ARHGAP44, ASNA1, ATP6V0E1, ATP6V1G2, BACE2, BECN1, BLVRA, BSN, C9orf3, CACNA2D2, CAMKK1, CAPNS1, CAV1, CCP110, CCT6A, CDK2AP1, CEND1, CFL1, CHST14, CKAP2L, CMTM6, CNPY2, COL3A1, CORO6, COX6B1, CPNE9, CRTAP, CSK, CSPG4, CTNNA3, DCTPP1, DDOST, DNAJC6, DOC2A, DOCK9, DUSP6, DUSP8, EFNA4, EIF4B, EIF6, EPN2, F2R, FAM129B, FAM136A, FAM57B, GALE, GAS2L3, GCM2, GGCT, GPR37L1, GSDMD, GUSB, GYPC, HDAC1, HDAC4, HIST1H2AB, HIST1H2AD, HIST1H2BK, HIST1H3D, HIST1H4L, HNRNPF, ICA1L, IGLL5, IL17RB, ILF2, ITGA10, ITGB3, ITPRID1, ITPRIPL2, ITSN1, KCNIP4, KCNJ11, KIAA1958, KLF10, KPNA2, LAMB2, LDHA, LMAN1L, MAD2L1, MAP3K1, MAPK9, MAPRE1, MEA1, MGP, MKI67, MLEC, MSN, MYL12B, NACA, NAPB, NCOA1, NEU1, NME1-NME2, NRIP2, NTSR2, PABPC1, PAGE5, PAX6, PCGF1, PHYHIP, PITPNC1, PLEKHM3, PLEKHO2, PLOD3, PMAIP1, PPFIA4, PREPL, PSMA1, PSMB3, PSMB4, PTMA, PYGL, RAB13, RASD1, RET, RHOA, RIMS3, RPL13A, RPL35A, RPS17, RPS9, RUNDC3A, RUNX3, S100A4, SCAMP5, SEC11A, SLC12A5, SLC25A18, SLC26A2, SLC43A3, SLC4A2, SLC8A2, SLCO3A1, SNRPB, SNTG2, SNX22, SPAG9, SPG21, ST18, ST6GALNAC2, SUPT4H1, SURF1, SYDE1, SYNE4, TAS2R19, TBC1D30, TEAD2, TGFBI, TGIF2, TJP3, TMEM116, TMSB10, TPI1, TPM3, TRIM63, TRMT11, TRMT44, TTC32, TUBA4A, TUBB, TUBB6, TXNDC12, UBL5, VAMP1, VAT1, VIPR1, VWA5B2, WASF1, WDR38, WEE1, YPEL4, YTHDF2, ZBTB42, ZDHHC11, ZFHX2, ZNF217, ZNF337, ZNF385B, ZNF488, ZNF708, ZWINT.
Evaluated genes: AAR2, ACTL6A, AEBP1, ANXA2, ATP6V0E1, BACE2, CAV1, CDK2AP1, CHST14, CMTM6, COL3A1, CRTAP, CSK, CSPG4, EFNA4, EIF4B, F2R, FAM129B, GAS2L3, GSDMD, GUSB, GYPC, HDAC1, HIST1H2AB, HIST1H2AD, HIST1H2BK, HIST1H3D, HIST1H4L, HNRNPF, IGLL5, ITGA10, ITGB3, ITPRIPL2, LAMB2, MAP3K1, MAPRE1, MGP, MKI67, MLEC, MSN, NACA, NEU1, PABPC1, PAGE5, PCGF1, PLEKHO2, PLOD3, PMAIP1, PTMA, RAB13, RHOA, RPL13A, RPL35A, RPS17, RPS9, RUNX3, S100A4, SEC11A, SLC26A2, SLC43A3, SLC4A2, SPG21, ST6GALNAC2, SURF1, SYDE1, TEAD2, TGFBI, TGIF2, TRIM63, TUBB6, VAT1, WDR38, WEE1, ZBTB42, ZNF217, ACBD7, AP3B2, ARHGAP44, ATP6V1G2, BSN, CACNA2D2, CAMKK1, CEND1, CORO6, CPNE9, DNAJC6, DOC2A, GCM2, ICA1L, ITPRID1, KCNIP4, MAPK9, NAPB, NTSR2, PHYHIP, PLEKHM3, PPFIA4, PREPL, RASD1, RET, RIMS3, RUNDC3A, SCAMP5, SLC12A5, SLC8A2, SYNE4, TBC1D30, TRMT11, TTC32, TUBA4A, VAMP1, VIPR1, VWA5B2, WASF1, YPEL4, ZNF385B, ZNF488, ZNF708.
Evaluated genes: ACLY, ACTG1, AP2S1, APEH, ARF1, ASNA1, BECN1, BLVRA, CAPNS1, CCT6A, CFL1, CKAP2L, CNPY2, COX6B1, CRTAP, DCTPP1, DDOST, DUSP6, EIF6, FAM136A, GALE, GGCT, ILF2, KLF10, KPNA2, LDHA, MAD2L1, MEA1, MYL12B, NEU1, NME1-NME2, PSMA1, PSMB3, PSMB4, PYGL, SNRPB, SNX22, SUPT4H1, TMSB10, TPI1, TPM3, TUBB, TXNDC12, UBL5, YTHDF2, ZWINT, A2ML1, ACSBG2, AIFM3, C9orf3, CCP110, CTNNA3, DOCK9, DUSP8, EPN2, FAM57B, GPR37L1, HDAC4, IL17RB, ITSN1, KCNJ11, KIAA1958, LMAN1L, NCOA1, NRIP2, PAX6, PITPNC1, RASD1, SLC25A18, SLCO3A1, SNTG2, SPAG9, ST18, TAS2R19, TJP3, TMEM116, TRMT44, ZDHHC11, ZFHX2, ZNF337.
Evaluated genes: AAR2, ACTL6A, AEBP1, ANXA2, ATP6V0E1, BACE2, CAV1, CDK2AP1, CHST14, CMTM6, COL3A1, CRTAP, CSK, CSPG4, EFNA4, EIF4B, F2R, FAM129B, GAS2L3, GSDMD, GUSB, GYPC, HDAC1, HIST1H2AB, HIST1H2AD, HIST1H2BK, HIST1H3D, HIST1H4L, HNRNPF, IGLL5, ITGA10, ITGB3, ITPRIPL2, LAMB2, MAP3K1, MAPRE1, MGP, MKI67, MLEC, MSN, NACA, NEU1, PABPC1, PAGE5, PCGF1, PLEKHO2, PLOD3, PMAIP1, PTMA, RAB13, RHOA, RPL13A, RPL35A, RPS17, RPS9, RUNX3, S100A4, SEC11A, SLC26A2, SLC43A3, SLC4A2, SPG21, ST6GALNAC2, SURF1, SYDE1, TEAD2, TGFBI, TGIF2, TRIM63, TUBB6, VAT1, WDR38, WEE1, ZBTB42, ZNF217.
Evaluated genes: ACBD7, AP3B2, ARHGAP44, ATP6V1G2, BSN, CACNA2D2, CAMKK1, CEND1, CORO6, CPNE9, DNAJC6, DOC2A, GCM2, ICA1L, ITPRID1, KCNIP4, MAPK9, NAPB, NTSR2, PHYHIP, PLEKHM3, PPFIA4, PREPL, RASD1, RET, RIMS3, RUNDC3A, SCAMP5, SLC12A5, SLC8A2, SYNE4, TBC1D30, TRMT11, TTC32, TUBA4A, VAMP1, VIPR1, VWA5B2, WASF1, YPEL4, ZNF385B, ZNF488, ZNF708.
Evaluated genes: ACLY, ACTG1, AP2S1, APEH, ARF1, ASNA1, BECN1, BLVRA, CAPNS1, CCT6A, CFL1, CKAP2L, CNPY2, COX6B1, CRTAP, DCTPP1, DDOST, DUSP6, EIF6, FAM136A, GALE, GGCT, ILF2, KLF10, KPNA2, LDHA, MAD2L1, MEA1, MYL12B, NEU1, NME1-NME2, PSMA1, PSMB3, PSMB4, PYGL, SNRPB, SNX22, SUPT4H1, TMSB10, TPI1, TPM3, TUBB, TXNDC12, UBL5, YTHDF2, ZWINT
Evaluated genes: A2ML1, ACSBG2, AIFM3, C9orf3, CCP110, CTNNA3, DOCK9, DUSP8, EPN2, FAM57B, GPR37L1, HDAC4, IL17RB, ITSN1, KCNJ11, KIAA1958, LMAN1L, NCOA1, NRIP2, PAX6, PITPNC1, RASD1, SLC25A18, SLCO3A1, SNTG2, SPAG9, ST18, TAS2R19, TJP3, TMEM116, TRMT44, ZDHHC11, ZFHX2, ZNF337
No functional results have been found significant.
Evaluated genes: ACTL6A, AEBP1, ATP6V0E1, BACE2, CAV1, CDK2AP1, CHST14, COL3A1, CSK, CSPG4, FAM129B, GAS2L3, GSDMD, HDAC1, HIST1H3D, HIST1H4L, ITGA10, ITGB3, ITPRIPL2, LAMB2, MAP3K1, MAPRE1, MLEC, MSN, PABPC1, PAGE5, PLEKHO2, PLOD3, PMAIP1, PTMA, RHOA, SEC11A, SLC26A2, SLC43A3, SURF1, SYDE1, TEAD2, TGIF2, VAT1, WDR38, ZBTB42, ZNF217, AP3B2, ATP6V1G2, BSN, CEND1, CORO6, CPNE9, DOC2A, ICA1L, KCNIP4, MAPK9, PHYHIP, PPFIA4, RASD1, RUNDC3A, SCAMP5, SLC12A5, SLC8A2, TUBA4A, VAMP1, VIPR1, VWA5B2, WASF1, YPEL4, ZNF385B, ACLY, ACTG1, AP2S1, ARF1, CAPNS1, CCT6A, COX6B1, DCTPP1, DUSP6, GALE, GGCT, KLF10, LDHA, MEA1, MYL12B, NEU1, NME1-NME2, PSMB3, PSMB4, SNRPB, SUPT4H1, TMSB10, TPI1, UBL5, YTHDF2, A2ML1, ACSBG2, AIFM3, C9orf3, CTNNA3, DUSP8, EPN2, GPR37L1, IL17RB, ITSN1, KIAA1958, SLC25A18, SPAG9, ST18, TJP3.
Evaluated genes: ACTL6A, AEBP1, ANXA2, CDK2AP1, CMTM6, CRTAP, F2R, GYPC, HNRNPF, MKI67, PABPC1, RAB13, SPG21, ST6GALNAC2, TGFBI, TUBB6, WEE1, ATP6V1G2, BSN, CAMKK1, NAPB, PLEKHM3, PPFIA4, SCAMP5, SLC12A5, WASF1, YPEL4, ASNA1, CNPY2, TPI1, TXNDC12, A2ML1, IL17RB, LMAN1L, PAX6, PITPNC1, SPAG9, TAS2R19, TMEM116, TRMT44.
No functional results have been found significant.
Evaluated genes: AAR2, CSPG4, EFNA4, EIF4B, GUSB, GYPC, HIST1H2AB, HIST1H2AD, HIST1H2BK, NACA, PLOD3, PTMA, RAB13, RPL13A, RPL35A, RPS17, RPS9, RUNX3, S100A4, SLC4A2, SYDE1, ARHGAP44, ATP6V1G2, CACNA2D2, DNAJC6, MAPK9, PREPL, RET, RIMS3, TBC1D30, TRMT11, TUBA4A, APEH, BECN1, BLVRA, CFL1, CNPY2, DCTPP1, DDOST, EIF6, FAM136A, GGCT, ILF2, KPNA2, MAD2L1, MYL12B, PSMA1, PYGL, TPM3, TUBB, ZWINT, C9orf3, CCP110, DOCK9, HDAC4, NRIP2, PAX6, SLCO3A1, SNTG2, ST18, ZDHHC11, ZFHX2, ZNF337.