Functional profiling
Key concepts
Assessed profiles:
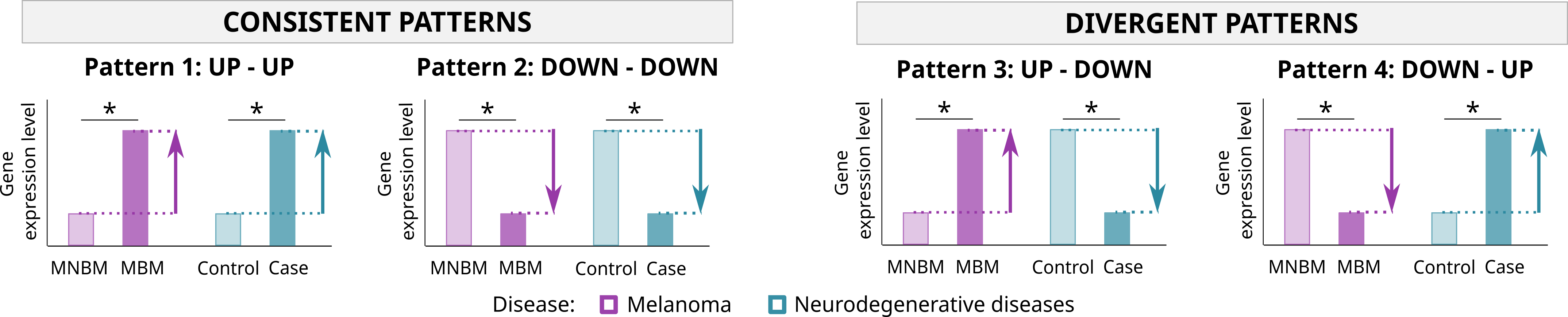
- By pattern: functional analysis of genes presenting the same dysregulation pattern in MBM-1-2 and in neurodegenerative diseases. Patterns evaluated:
- By neurodegenerative disease: functional analysis of genes altered in MBM-1-2 and a specific neurodegenerative disease, independently of their dysregulation pattern. Neurodegenerative diseases evaluated:
- Cortex region of Alzheimer’s disease (AD-CT)
- Hippocampus region of Alzheimer’s disease (AD-HP)
- Substantia nigra region of Parkinson’s disease (PD-SN)
Functional results
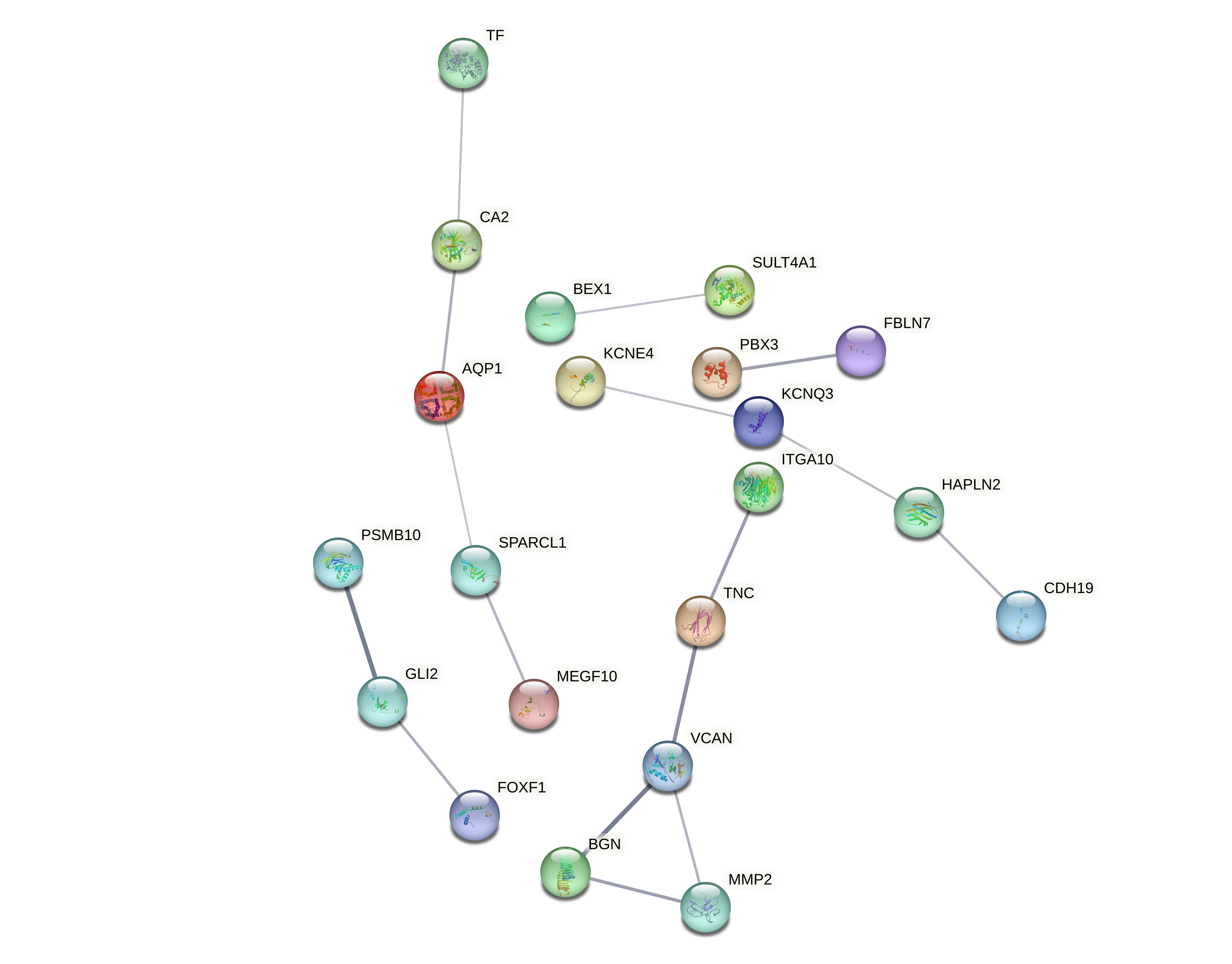
Evaluated genes: AQP1, ATRNL1, BEX1, BGN, CA2, CD48, CDH19, CNTD1, DLL1, DNAJC6, DPYSL3, EPSTI1, FAM107B, FBLN7, FILIP1L, FMNL2, FOXF1, GLI2, HAPLN2, ITGA10, KCNE4, KCNQ3, KLHL8, LBH, MEGF10, MKLN1, MMP2, MORN4, NAV2, NFIB, OR7A5, PALLD, PARP15, PBX1, PBX3, PMP2, PRDX6, PSMB10, SEPTIN6, SLC13A4, SLTM, SPARCL1, ST6GALNAC3, STXBP5L, SULT4A1, TF, TNC, UCHL1, VCAN, ZFP36L1, ZIC4, ZNF521, ZNF536.
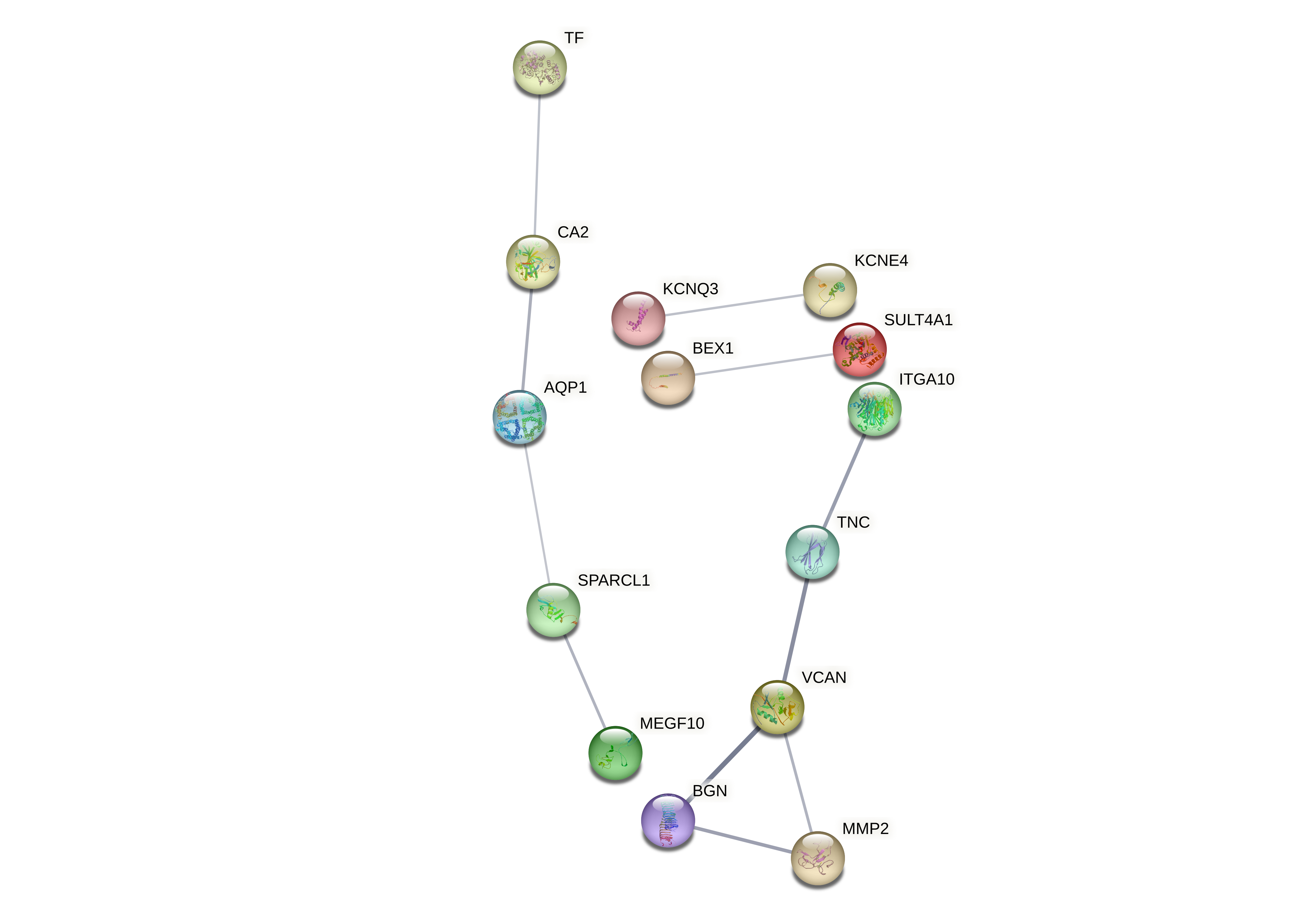
Evaluated genes: CA2, CDH19, CNTD1, FMNL2, HAPLN2, ITGA10, KLHL8, MEGF10, MKLN1, NAV2, OR7A5, PMP2, PRDX6, SLC13A4, ST6GALNAC3, TF, ZIC4, DPYSL3, EPSTI1, FBLN7, FILIP1L, LBH, PBX1, PSMB10.
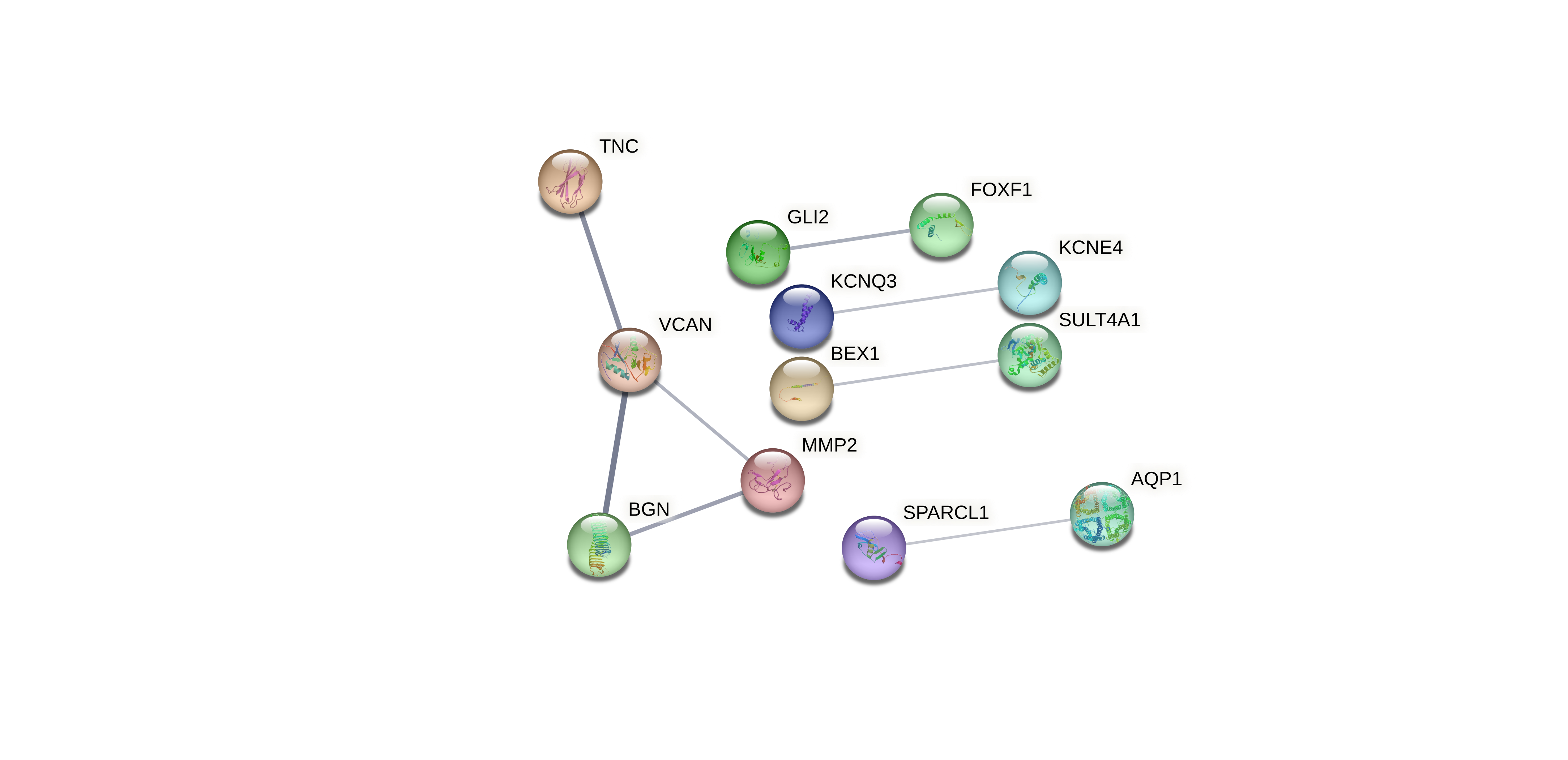
Evaluated genes: ATRNL1, BEX1, DNAJC6, KCNQ3, MORN4, SPARCL1, STXBP5L, SULT4A1, UCHL1, ZNF536, AQP1, BGN, CD48, DLL1, DPYSL3, FAM107B, FOXF1, GLI2, KCNE4, MMP2, NFIB, PALLD, PARP15, PBX3, SEPTIN6, SLTM, TNC, VCAN, ZFP36L1, ZNF521.
Evaluated genes: CA2, CDH19, CNTD1, FMNL2, HAPLN2, ITGA10, KLHL8, MEGF10, MKLN1, NAV2, OR7A5, PMP2, PRDX6, SLC13A4, ST6GALNAC3, TF, ZIC4.
No functional results have been found significant.
Evaluated genes: DPYSL3, EPSTI1, FBLN7, FILIP1L, LBH, PBX1, PSMB10.
Few genes for performing functional analyses.
Evaluated genes: ATRNL1, BEX1, DNAJC6, KCNQ3, MORN4, SPARCL1, STXBP5L, SULT4A1, UCHL1, ZNF536.
No functional results have been found significant.
Evaluated genes: AQP1, ATRNL1, BEX1, BGN, CA2, CDH19, DLL1, DPYSL3, FAM107B, FILIP1L, FMNL2, FOXF1, ITGA10, KCNE4, KCNQ3, MEGF10, MMP2, MORN4, NAV2, NFIB, PALLD, PBX3, PMP2, PRDX6, SLC13A4, SLTM, SPARCL1, ST6GALNAC3, SULT4A1, TF, TNC, UCHL1, VCAN, ZFP36L1.
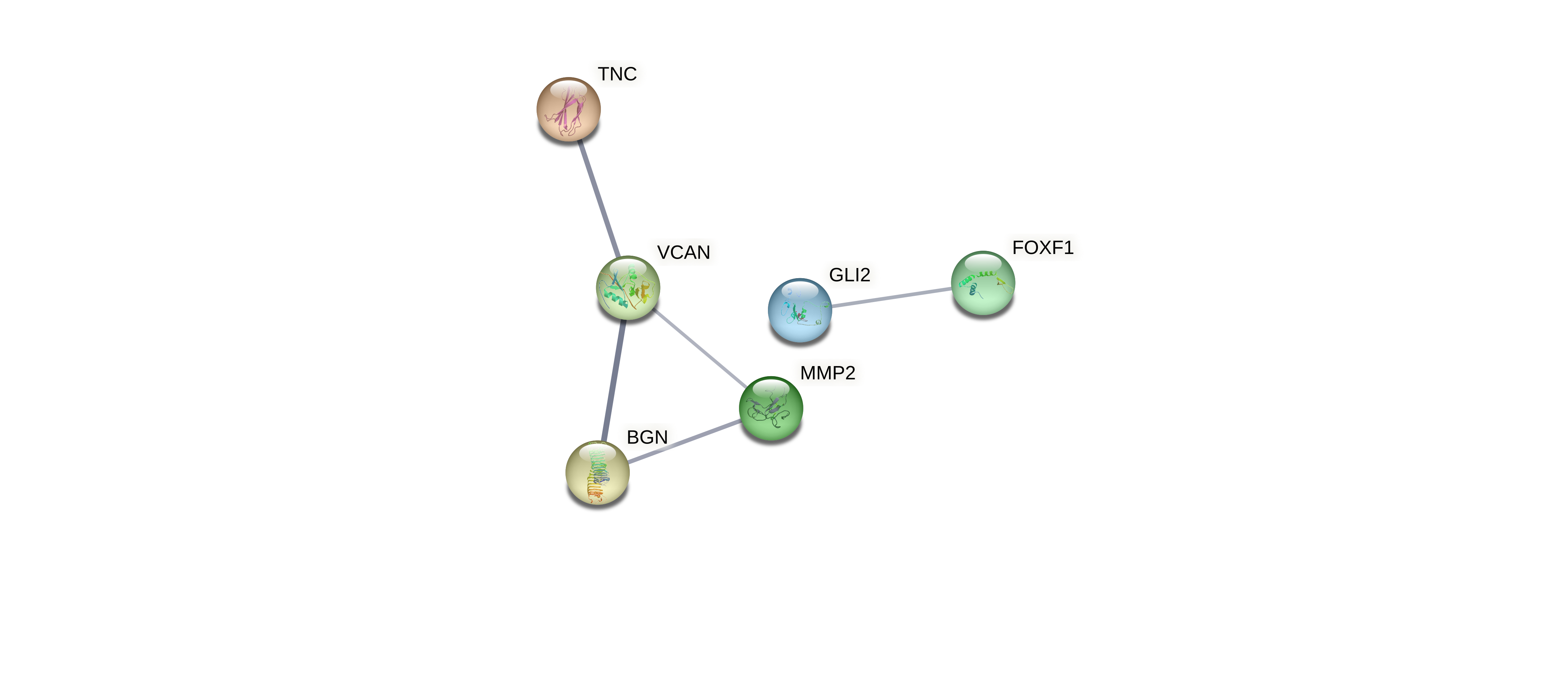
Evaluated genes: BEX1, CD48, CNTD1, EPSTI1, GLI2, KLHL8, MKLN1, NFIB, PALLD, PARP15, PBX3, PMP2, PSMB10, ST6GALNAC3, UCHL1, VCAN, ZIC4, ZNF521.