This is an old revision of the document!
Input Parameters
Go to Network Miner section: Babelomis > Functional Analysis > Set Enrichment Analysis > Network Miner
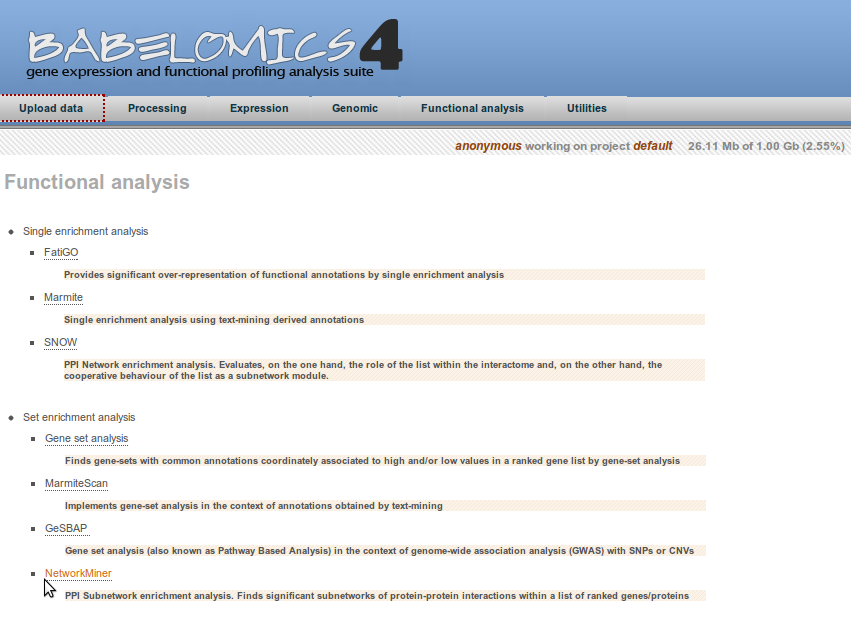
In the Network Miner tool you will find the following options:
1. Select your data (mandatory)
Browse your own ranked list. The ranked list must be a plain text file and should contain with two columns. The first column must be numeric. The second column must contain your genes, transcripts or proteins. Alternatively, you can upload a plain text file with just one column containing your genes, transcripts or proteins. If a second column file is provided, then you can tell Network Miner how to rank your list. If not, the default order will be considered. See an input example file: example.txt.
2. Select your seed data (optional)
Browse your seed list. The seed list must be a plain text file with one column. This column must contain your genes, transcripts or proteins.
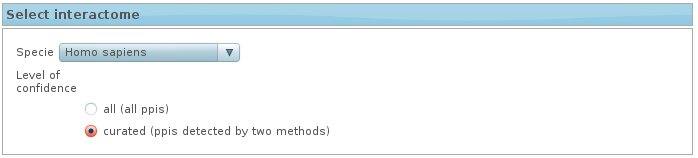
3. Select interactome
Choose the interactome you want to use in the analysis.
- Select the specie
- Select the level of ppis confidence:
- non-curated interactome (all ppis)
- curated interactome (ppis detected by at least two methods).
See About Interactomes databases for more information about their sources, statistics and generation.
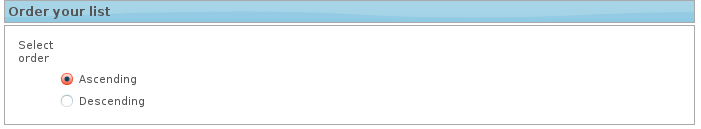
4. Order your list
When a two columns file is provided it is possible to sort the list by the second column containing the experimental parameter. For example, if you have upload a two columns file in which the first column contains the transcripts and the second column contains the statistic value from differential expression analysis, therefore by choosing descending, Network Miner will prioritize the transcripts with the highest statistic value (you will analyse the genes up regulated) whereas by choosing ascending Network Miner will prioritize the transcripts with the lowest statistic value (you will analyse the genes down regulated).
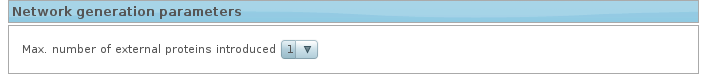
5. Network generation parameters
MCN is generated calculating the shortest paths among all the pairs of proteins/genes in your list. The analysis can be performed by adding one intermediate node (genes originally not included in the list that can actually link two or more genes of the list), so this option is to indicate if an external gene could be introduced to generate the MCN. Including external genes makes the approach more robust to noise and also overcomes some intrinsic limitations of experimental designs based in differential expression, such as the impossibility of detecting nodes whose expression is not different across the conditions compared.
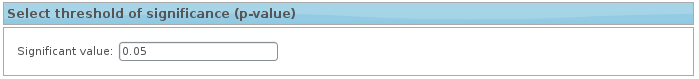
6. Select threshold of significance (p-value)
You could select threshold of significance to perform the Network Miner analysis.
7. Job information
Put a job name and run the analysis. Jobs may last approximately less than a minute but the time may vary depending on the characteristics of the analysis.