Table of Contents
Data description
Working plan
We have several candidate profiles of mutations because it is clear the kind of mutation we are searching but sometimes we do not have a specific clues.
How many variants do you detect for each scenario?
A. Individual filters
- Total number of variants.
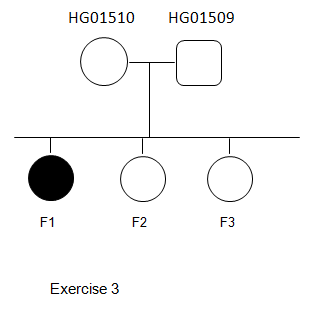
- Recessive inheritance
- Dominant inheritance (mother is affected).
- For these region, only chromosome: 3,19
- Evaluate the same regions, including start and end at the first region: 3:1000-200000,19
- For this gene: SDC3
- Description of variants. How many SNVs, INDELs, MNVs, SVs, CNVs?
- Variants with MAF (Minimum Allelic Frequency) < 0.05 for AFR populations in 1000 Genomes phase 3
- Variants with MAF (Minimum Allelic Frequency) < 0.05 for African American population in ESP 6500
- Variants with MAF (Minimum Allelic Frequency) < 0.05 for EXAC population in ESP 6500
- For SIFT <0.05 and polyphen >0.95, because it is likely pathogenic
B. Progressive selection
- Recessive inheritance + variants in candidate genes related to “stress fiber” function. (Clue: Gene Ontology filter).
- Another scenario:
- Retrieve the filter for recessive inheritance (small icon).
- Variants in candidate genes related to “calcium ion binding” function. (Clue: Gene Ontology filter).